Jack Heuer ’24: Lab of Medical Imaging and Computation at Massachusetts General Hospital
- The Rivers School

- Oct 12, 2023
- 4 min read
In the past few years, I have used computer science to solve problems such as creating a game of checkers or sorting a list, but I’ve never created something that could help a real person in the world. I was extremely excited to earn an internship with the Lab of Medical Imaging and Computation (LMIC) because I could finally make something useful. The LMIC works to create machine learning models to help advance the world of medical imaging and provide accessible and efficient medical care for the world.

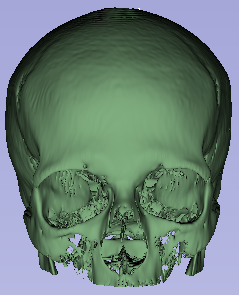
I began my internship learning how to navigate the 3D slicer tool. With this tool, I was able to work with CT scans in order to segment specific parts of the body. Creating a threshold of density units known as HUs, I was able to segment the skull from a head CT scan and the lungs from a chest CT scan. Next, we learned how to write code in Python that would semi-automatically segment these parts of the body without the need for the 3D slicer tool. This program would use the same density units and a premade library in order to draw borders around what was important and create a mask. With these masks, we were able to delete certain parts of each layer in a CT scan and only leave the parts of the scan that included the body parts we were looking for.
Slices of a brain CT scan with coordinating masks 3D model formed with stacked segmented masks
Next, we were tasked with using the Unet model, a popular type of convolutional neural network, in order to create a machine-learning model that would automatically segment thousands of lungs at once. After finding data online, I standardized the photos by rotating them and creating maximum pixel values for each chest X-ray. I then trained the model on this new data and got an accuracy rate of 97%. I learned new things which I employed in my model such as how to reduce memory cost and how to slow learning rates so as to not overtrain my model. With these generated masks I was then able to cut just the lung areas out of the X-ray and be left with a lung image.

Original x-ray image | Lung segmentation mask | Original image cut by the mask’s boundaries to the right
We were then tasked with creating a model that would predict whether or not the lungs in an X-ray were normal or abnormal with some type of affliction. Using the prior model’s generated masks, I found the top points and bottom points and linked them in order to create a mask of the entire chest area. With these new masks, I cut the original images again and used these new cut images in order to train my diagnosis model.

Cutting out the unimportant parts of each X-ray helped to eliminate a confounding effect that can happen when the model starts to look at the little letters in each photo which are used for labeling. Without cutting out the region of interest the model might look at the letters and create a prediction based on that instead of the image itself.
With my edited X-ray images I was able to predict a diagnosis correctly 95% of the time. While this number might seem high it would be an issue for 5% of the patients to receive an incorrect diagnosis and end up treating an ailment that didn’t actually exist or be left with a false sense of security.
My final project was a group project with the other three interns. We wanted to create an application that would crawl the internet and download all the images we possibly could, classify these images as medical images (such as x-rays, CT scans, ultrasounds, and pet scans) or not medical images, sort these images by type of medical image, sort each type of medical image by part of the body if it was possible, and finally create a website where anyone could download these images for free.

Medical images are commonly withheld from the public because there are a lot of patient confidentiality rules that the hospitals do not want to break. Sites like kaggle have only a limited amount of data for medical machine learning. With many people wanting to help create these applications for medical imaging but only a limited amount of data it provides a problem. More data means more and better models which means more people helped around the world.
One part of the project I completed was a medical vs. non-medical classifier. I used lots of skills I learned throughout the summer and was able to achieve an accuracy of 99.5% which I was happy with.

I also learned how to use Scrapy, a popular crawling tool in order to create a crawler that would download all the medical images from the image library Flickr. With all the models created and ready we are now working on hosting a website off of AWS (Amazon Web Services).

This summer, I learned a lot of Python and machine learning tools but more importantly, I learned how to better collaborate with a team in a professional environment. Working with different types of people can be very helpful in obtaining different perspectives that help to see problems from different angles. As we work to finish our final project together I will continue to employ the skills I have learned.
I am extremely grateful to have been surrounded by friendly people who supported me with tactful advice and an inviting environment. Seeing people doing interesting and meaningful work in the computer science industry has grown my interest exponentially. Thank you to Dr. Synho Do for hosting this internship and creating an unforgettable experience for us interns. Also thanks to Mr. Schlenker for providing me with this opportunity.






Main di kabar4d selalu gacor, langsung WD bro!